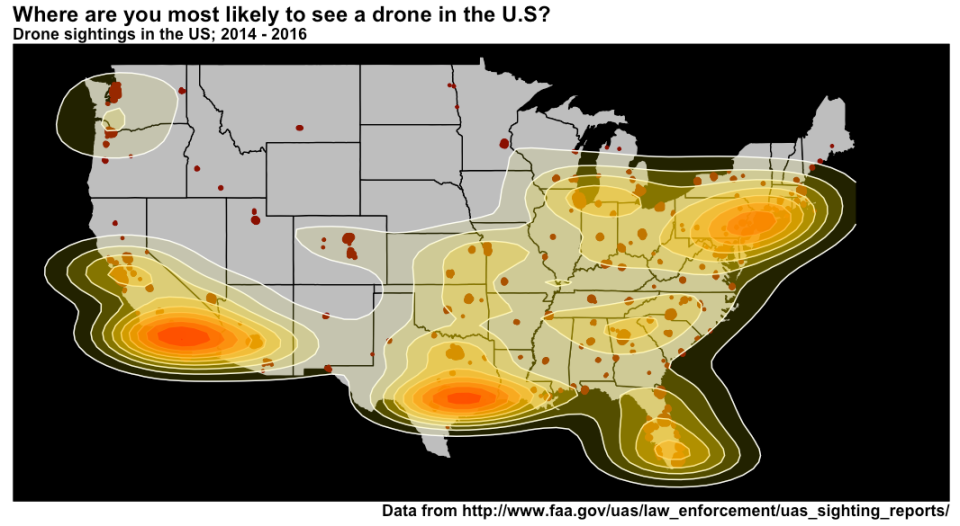
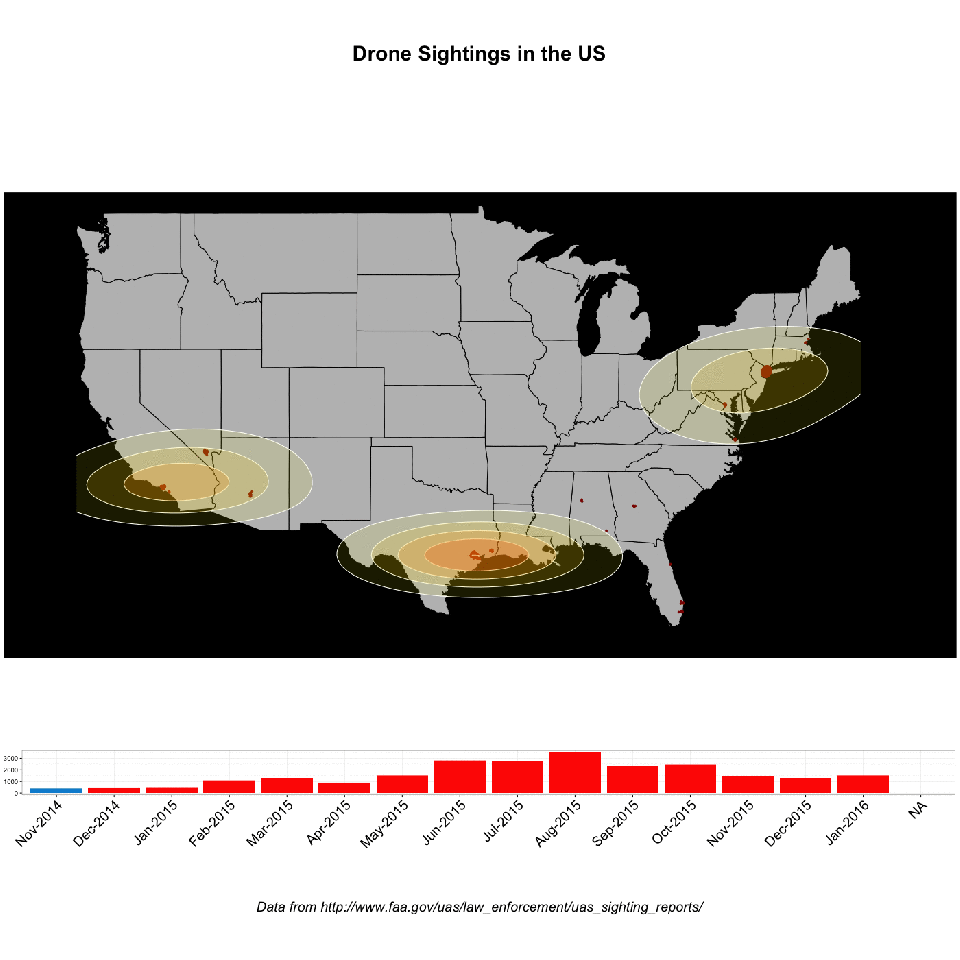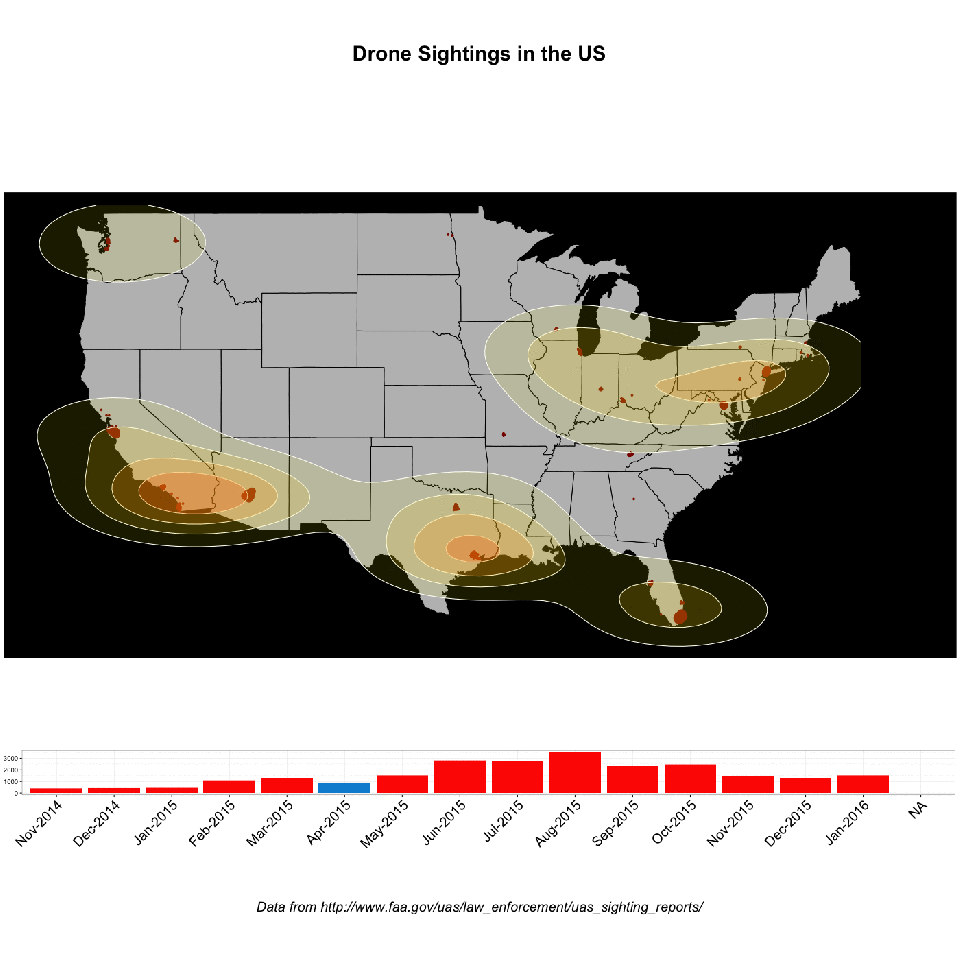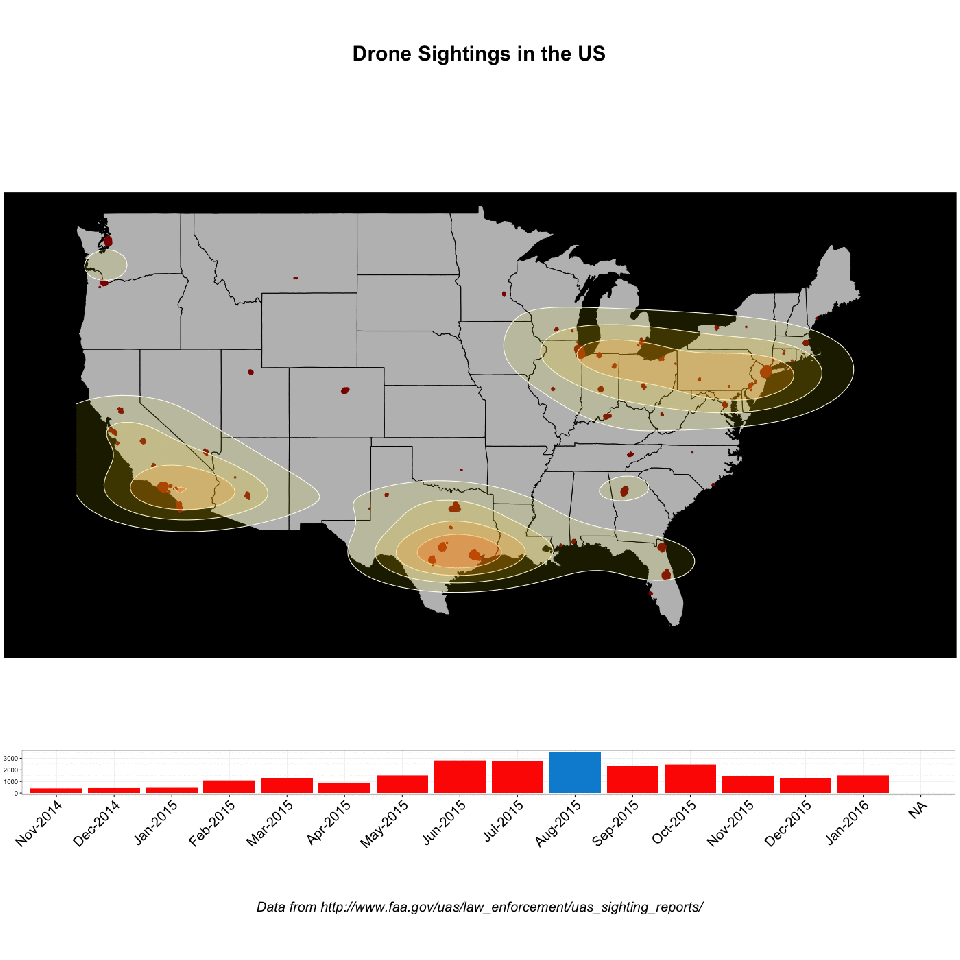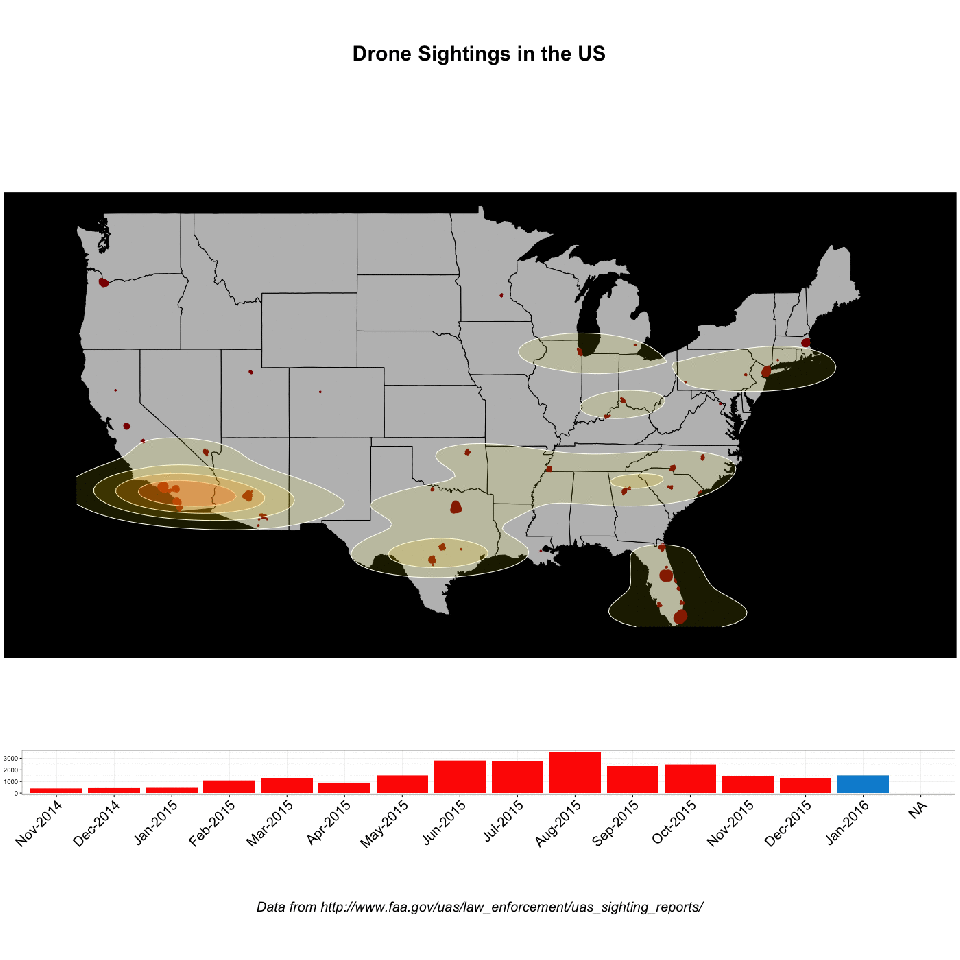
Record linkage is an immensely useful, albeit tedious, task. Easily, one of the most common requests I’ve encountered in an enterprise environment is “wouldn’t it be great to see all of this data from all these different places in the same location?” (AND make it make it predictive, make it interactive, make it into a web app and why doesn’t it look like Microsoft Powerpoint?, and update me on progress every 3 days…). Never mind that these two databases might as well have been developed on different planets: no hard matching key, no consistent data hierarchy, sometimes the only unique identifier is a row number!
Your team: Probably just you, but here are the hats you’ll have to wear:
– Business owner: Defines the problem. Identifies and interviews stakeholders, asks “what would be useful for you and/or where can I get that data?”
– Extraction Team: Works with dev ops / IT of the databases in question to automate access and set up a centralized repository (even if that’s just a local desktop)
– Transformation Team: Ideally, 1 person per database you wish to link. Owns the SQL queries for rolling up data and works with stakeholders to verify the rowcounts are mostly correct (most of the time, nobody ever knows if the rowcounts are exactly correct)
– Linkage modeler: Creates the linkage table. Needs strong data munging skills and should know how to build a classifier
– Reporting Team: researches industry reports, creates top-down list of desired views and works to reconcile with available data
– Visualization Team: because, why do anything if it’s not fun?
Planning and research
If you’re using R, the packages available for this specific problem are somewhat sparse. You have agrep() and adist() for linkage, which both help, but the real problem is one of computation. Consider 12,000 possible names in one database and 115,000 in the other, that’s over a billion possible matches. In addition, how do you define what a match is? How many false positives are you willing to tolerate? See here for some ideas: http://stackoverflow.com/questions/6683380/techniques-for-finding-near-duplicate-records
The RecordLinkage package, which for some reason was archived and abandoned, offers some excellent tools for dealing with the computation issue. The package is the result of work by Murat Sariyar and Andreas Borg, and they lay out some key concepts in this article: http://journal.r-project.org/archive/2010-2/RJournal_2010-2_Sariyar+Borg.pdf
Note: You’ll need to install RecordLinkage from the archive
Key concepts:
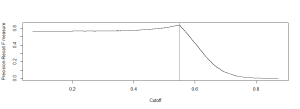
– Blocking: the concept of only creating possible match pairs based on some hard criterion, such as only searching for pairs within the group where the first letter of the first word matches. Drastically reduces computation time and effort, and actually increases accuracy (or, more technically, increases precision at the expense of recall, i.e., failing to identify true positives).
– Phonetic functions (e.g., soundex) and string comparison (e.g., levenshein distance): Functions for comparing like strings. Useful if using RecordLinkage for weighting, and can also be very useful when used in conjunction with blocking (i.e., compare this string to all other strings which sound like it)
Before we get to the code, some key learnings:
– Pre-processing is IMMENSELY valuable in this process. Below, the functions CleanseNames(), SplitNames(), StandardizeWord(), MapFrequency(), and RearrangeWord() are the heart of the pre-processing and cleansing, and are responsible for 80-90% of the matches.
– The ApplyAdist() and ModifyAdist() functions at the bottom can be used to judge relative distances between two character strings
Finally, performance:
The below code, while heavily obfuscated, achieved an identical match rate to the commercially available D+B service which performs the same task.
#============================================================================
#============================================================================
#================= ENTERPRISE RECORD LINKAGE v.3 ==============================
#============================================================================
#============================================================================
#============================================================================
#============================================================================
# This is my third attempt at the linkage table creation process
# -Tim Kiely
# 8/10/2014
# timothy.j.kiely@gmail.com
#============================================================================
libs <- c("RODBC", "utils", "tm", "stringr", "gdata", "dplyr", "lubridate","beepr")
lapply (libs, library, character.only = TRUE, logical.return=TRUE)
#make sure you've already installed RecodLinkage from the archive
library(RecordLinkage)
#============================================================================
#============================================================================
#===================Pre-Processing for Record Linkage========================
#============================================================================
#============================================================================
#
# First, for both dataframes you wish to merge, create a character string variable called "Company.Name.Compress" which contains names of the companies
# Note: this step cleanses "Company.Name.Compress"
# Also replaces common words, e.g. "INCORPORATION" replaces "INC." and "Inc"
#
#============================================================================
CleanseNames <- function(companies) {
##Remove Special Chars
companies$Company.Name.Compress <- gsub("\\.", "", companies$Company.Name.Compress)
companies$Company.Name.Compress <- gsub("'", "", companies$Company.Name.Compress)
companies$Company.Name.Compress <- gsub("\\ &", " AND ", companies$Company.Name.Compress)
## Convert to UPPER CASE
companies$Company.Name.Compress <- toupper(companies$Company.Name.Compress)
## Remove Stopwords #NOTE: REMOVED THIS STEP: REMOVES IMPORTANT IDENTIFYING INFO AND HINDERS MATCH ABILITY
#companies$Company.Name.Compress <- removeWords(companies$Company.Name.Compress, stopwords("english"))
## Remove Punctuations
companies$Company.Name.Compress <- removePunctuation(companies$Company.Name.Compress)
## Eliminating Extra White Spaces
companies$Company.Name.Compress <- stripWhitespace(companies$Company.Name.Compress)
## Eliminate leading White Space
companies$Company.Name.Compress <- gsub("^ ", "",companies$Company.Name.Compress)
## Eliminate trailing White Space
companies$Company.Name.Compress <- gsub(" $", "", companies$Company.Name.Compress)
## Remove Numbers
#companies$Company.Name.Compress <- removeNumbers(companies$Company.Name.Compress)
return(companies)
}
df.companies.cleansed <- CleanseNames(df.companies)
df.db.companies.cleansed <- CleanseNames(df.db.companies)
#============================================================================
#============================================================================
#========================= WORD REPLACEMENT =================================
#============================================================================
#============================================================================
#
# Splits the company names and stores the first 5 words in seperate variables.
# If the name has less then 5 words then the remaining words are
# automatically assigned NA
#
# Note: this is the most time-consuming preprocessing operation
# (excluding record linkage, which is computationally expensive)
#
# If optimization is desired, this function could be improved
#
#============================================================================
SplitNames <- function(companies){
companies <- data.frame(companies
,First.Word= NA
,Second.Word= NA
,Third.Word= NA
,Fourth.Word= NA
,Fifth.Word= NA
,stringsAsFactors=F)
companies <- companies %.% group_by(1:n()) %.%
mutate(LinLen = length(unlist(strsplit(Company.Name.Compress," ")))
,First.Word = ifelse(LinLen<1,"",toupper(word(Company.Name.Compress,1)))
,Second.Word = ifelse(LinLen<2,"",toupper(word(Company.Name.Compress,2)))
,Third.Word = ifelse(LinLen<3,"",toupper(word(Company.Name.Compress,3)))
,Fourth.Word = ifelse(LinLen<4,"",toupper(word(Company.Name.Compress,4)))
,Fifth.Word = ifelse(LinLen<5,"",toupper(word(Company.Name.Compress,5)))
)
return(companies)
}
df.companies.split <- SplitNames(df.companies.cleansed)
df.db.companies.split <- SplitNames(df.db.companies.cleansed)
#============================================================================
#============================================================================
#========================== WORD STANDARDIZATION ============================
#============================================================================
#============================================================================
#
# Replaces Common interchangable words with standards, e.g.:
# "INCORPORATED" replaces "INC" and "Inc.", etc.
#
# Also, importantly creates the "Full.String" variable (concat of 5 words)
#
#============================================================================
StandardizeWord <- function (companies){
companies <- as.matrix(companies)
# Initial words are replaced by Final Words
initial <- c("CORP","USA","INC","CO","LTD","INTL","COMP","SVCS","SERVICE","HLDGS","MORTG","SYS","TECH",
"IND","SERV","FINL","LABS","TELECOMMUN","MTG","CP","PPTYS","SEC","COMMUN","DIST", "INCORPORATED",
"COS", "CORPO", "CORPOR", "CORPORA", "CORPORATI", "CORPORAT", "CORPORATIO", "CORPORATIONS",
"HLTHCR")
final <- c("CORPORATION","US","INCORPORATION","COMPANY","LIMITED","INTERNATIONAL","COMPANY","SERVICES",
"SERVICES","HOLDINGS","MORTGAGE","SYSTEM","TECHNOLOGY","INDUSTRIES","SERVICES", "FINANCIAL",
"LABORATORIES","TELECOMMUNICATIONS","MORTGAGE","CORPORATION","PROPERTIES","SECURITIES",
"COMMUNICATIONS","DISTRIBUTION", "INCORPORATION", "CORPORATION", "CORPORATION", "CORPORATION",
"CORPORATION", "CORPORATION", "CORPORATION", "CORPORATION", "CORPORATION", "HEALTHCARE")
word.standard <- as.matrix(cbind(initial, final)) # Creating a data.frame
# running the loop over the five words
var <- c("First.Word", "Second.Word", "Third.Word", "Fourth.Word", "Fifth.Word")
for(i in 1:5){
match.index <- match(companies[, var[i]], word.standard[,"initial"])
companies[, var[i]] <- ifelse(!is.na(match.index), word.standard[,"final"][match.index], companies[, var[i]])
}
# Creating a standardized full name again, by pasting back the five words
companies <- data.frame(companies, Full.String="NA")
companies <- companies %.%
group_by(1:n()) %.% mutate(Full.String=trim(paste(First.Word
, Second.Word
, Third.Word
, Fourth.Word
, Fifth.Word)
)
,Full.String=gsub("NA","",Full.String)
)
companies <- as.data.frame(companies,stringsAsFactors=F)
return(companies)
}
df2.companies <- StandardizeWord(df.companies.split)
df2.db.companies <- StandardizeWord(df.db.companies.split)
#============================================================================
#============================================================================
#========================== WORD SIGNIFICANCE CALC ==========================
#============================================================================
#============================================================================
#
# Calculate most significant word
# "INCORPORATED" replaces "INC" and "Inc.", etc.
#
# Also, importantly creates the "Full.String" variable (concat of 5 words)
#
#============================================================================
var <- c("First.Word","Second.Word","Third.Word","Fourth.Word","Fifth.Word")
words<-matrix()
db.words<-matrix()
words<-c(as.matrix(df2.companies[,var]))
words<-as.data.frame(words[!is.na(words)])
db.words<-c(as.matrix(df2.db.companies[,var]))
db.words<-as.data.frame(db.words[!is.na(db.words)])
names(words)="words"
names(db.words)="words"
words<-rbind(words,db.words)
freq.table<-words%.%group_by(words)%.%summarise(count=n())%.%arrange(-count)
freq.table <- as.matrix(freq.table)
# renaming the columns of frequency table
colnames(freq.table)[1] <- "Word"
colnames(freq.table)[2] <- "Freq"
MapFrequency <- function(companies, frequencies){
var <- c("First.Word", "Second.Word", "Third.Word", "Fourth.Word", "Fifth.Word")
# running the loop over 5 words
for(i in 1:5){
# renaming column name of "frequencies" to Freq.Word1, Freq.Word2 etc. in each iteration
colnames(frequencies)[2] <- paste("Freq.Word", i, sep="")
# merging Freq.Word1 for var[1] i.e. "First.Word" ; Freq.Word2 for var[2] i.e. "Second.Word" and so on..
companies <- as.matrix(merge(companies, frequencies, by.x=var[i], by.y="Word", all.x=TRUE))
}
companies <- as.data.frame(companies)
return(companies)
}
df2.companies <- MapFrequency(df2.companies,freq.table)
df2.db.companies <- MapFrequency(df2.db.companies,freq.table)
#=======================================================================================
# RearrangeWord
#
# Purpose: Rearrange the individual word columns and respective frequency columns as
# least frequent to most frequent from left to right. Word having the least
# frequency is most significant.
#
# Arguments:
# db.companies: A matrix having the individual word columns and the respective
# frequency columns
#
# Returns:
# A matrix with rearranged individual word columns according to their significance
#=======================================================================================
RearrangeWord <- function(db.companies){
as.matrix(db.companies)
var2 <- c("Freq.Word1", "Freq.Word2", "Freq.Word3", "Freq.Word4", "Freq.Word5")
var <- c("First.Word", "Second.Word", "Third.Word", "Fourth.Word", "Fifth.Word")
numeric.sorting <- function(temp) order(as.numeric(as.matrix(temp)))
# applying the sort function row wise
ordering.index <- apply(as.matrix(db.companies[, var2]), 1, numeric.sorting)
# correcting the ordering.index.
# It is actually a linear vector (not a matrix) and hence need to be corrected
ordering.index <- ordering.index + 5 * (col(ordering.index)-1)
# Rearranging the word frequencies
db.companies[,var2] <- t(matrix(t(db.companies[, var2])[ordering.index], nrow = 5, ncol = nrow(db.companies)))
# Rearranging the words themselves
db.companies[, var] <- t(matrix(t(db.companies[, var])[ordering.index], nrow = 5, ncol = nrow(db.companies)))
# Renaming the words
colnames(db.companies)[match(var, colnames(db.companies))] <- c("Sig.Word1", "Sig.Word2",
"Sig.Word3", "Sig.Word4", "Sig.Word5")
as.data.frame(db.companies)
return(db.companies)
}
df2.companies <- RearrangeWord(df2.companies)
df2.db.companies <- RearrangeWord(df2.db.companies)
#============================================================================
#============================================================================
#================ RECORD LINKAGE FEATURE SELECTION ==========================
#============================================================================
#============================================================================
#
# Re-Extracting the First Word, Second Word etc. features for improved blocking
df2.companies$Company.Name.Compress<-as.character(df2.companies$Company.Name.Compress)
df2.db.companies$Company.Name.Compress<-as.character(df2.db.companies$Company.Name.Compress)
df.companies.extra <- SplitNames(df2.companies)
df.db.companies.extra <- SplitNames(df2.db.companies)
#
df2.companies <- df.companies.extra
df2.db.companies <- df.db.companies.extra
#
#============================================================================
df2.companies$companies <- as.factor(df2.companies$companies)
df2.companies$Company.Name.Compress <- as.factor(df2.companies$Company.Name.Compress)
df2.companies$First.Word <- as.factor(df2.companies$First.Word)
df2.companies$Second.Word <- as.character(df2.companies$Second.Word)
df2.companies$Third.Word <- as.character(df2.companies$Third.Word)
df2.companies$Fourth.Word <- as.character(df2.companies$Fourth.Word)
df2.companies$Fifth.Word <- as.character(df2.companies$Fifth.Word)
df2.companies$Full.String <- as.character(df2.companies$Full.String)
df2.companies$LinLen <- as.integer(df2.companies$LinLen)
df2.companies$employer_key <- as.integer(df2.companies$employer_key)
df2.companies$Sig.Word1 <- as.factor(df2.companies$Sig.Word1)
df2.db.companies$db.companies <- as.factor(df2.db.companies$db.companies)
df2.db.companies$Company.Name.Compress <- as.factor(df2.db.companies$Company.Name.Compress)
df2.db.companies$First.Word <- as.factor(df2.db.companies$First.Word)
df2.db.companies$Second.Word <- as.character(df2.db.companies$Second.Word)
df2.db.companies$Third.Word <- as.character(df2.db.companies$Third.Word)
df2.db.companies$Fourth.Word <- as.character(df2.db.companies$Fourth.Word)
df2.db.companies$Fifth.Word <- as.character(df2.db.companies$Fifth.Word)
df2.db.companies$Full.String <- as.character(df2.db.companies$Full.String)
df2.db.companies$LinLen <- as.integer(df2.db.companies$LinLen)
df2.db.companies$Sig.Word1 <- as.factor(df2.db.companies$Sig.Word1)
df2.companies["X1.n.."]<-NULL
df2.companies["X1.n...1"]<-NULL
df2.companies["1:n()"]<-NULL
df2.db.companies["X1.n.."]<-NULL
df2.db.companies["X1.n...1"]<-NULL
df2.db.companies["1:n()"]<-NULL
df2.companies<-as.data.frame(df2.companies)
df2.db.companies<-as.data.frame(df2.db.companies)
table1<- df2.companies%.%select(companies=companies
,Company.Name.Compress
,First.Word
,Sig.Word1
,LinLen
,employer_key)
table2 <- df2.db.companies%.%select(companies=db.companies
,Company.Name.Compress
,First.Word
,Sig.Word1
,LinLen
)%.%
mutate(employer_key=as.integer(seq_along(companies)))
#============================================================================
#============================================================================
#================ RECORD LINKAGE: ===========================================
#================ MATCH PAIR GENERATION =================================
#============================================================================
#============================================================================
#
# Two additional functions for modifying and applying the base approximate distance function adist
# Modified verison "ModifyAdist" penalizes for incorrect first letter and increases penalty for substitutions (decreases chance of false positive)
ApplyAdist <- function(word1, word2){
tryCatch({
ged.string <- adist(word1, word2, counts = T,ignore.case=T)
# Assigning distinct weightages
ged <- sum(attr(ged.string, "counts"))
return(ged)
})
}
#
#
ModifyAdist <- function(word1, word2){
tryCatch({
ged.string <- adist(word1, word2, counts = T,ignore.case=T)
# Assigning distinct weightages
ged <- sum(attr(ged.string, "counts")* c(1, 1, 5))
# 1: Insertion 2: Deletion 3: Substitution
# Adding extra cost if the first character of the string differs
if(strsplit(attr(ged.string, "trafos"), split = "")[[1]][1] != "M") ged <- ged + 8
return(ged)
})
}
#
#
#
#============================================================================
#============================================================================
system.time({
# use this if you want to test on a subset:
tam.frac <- sample_frac(table1,1,replace=F)
db.frac <- sample_frac(table1,1,replace=T)
identity.tam <- as.numeric(tam.frac$employer_key)
identity.db <- as.numeric(db.frac$employer_key)
results <- RLBigDataLinkage(
dataset1 = tam.frac
,dataset2 = db.frac
,identity1 = identity.tam
,identity2 = identity.db
,blockfld = "First.Word"
,exclude = c("companies"
,"employer_key"
,"LinLen"
,"Sig.Word1"
,"First.Word")
,strcmp = "Company.Name.Compress"
,strcmpfun = "levenshtein"
#,phonetic = ""
#,phonfun = "pho_h"
)
})
results.fin <-
getPairs(
results
#,filter.match = c("match", "unknown", "nonmatch")
#,filter.link=c("link")
,max.weight = Inf
,min.weight = -Inf
#,withMatch=T
#,withClass=T
,single.rows=T
)
results.fin$Company.Name.Compress.1<-as.character(results.fin$Company.Name.Compress.1)
results.fin <- results.fin %.%
group_by(1:n()) %.%
mutate(NumChar = length(unlist(strsplit(Company.Name.Compress.1,""))))%.%
mutate(company_aDist = ApplyAdist(companies.1,companies.2)) %.%
mutate(company_MaDist = ModifyAdist(companies.1,companies.2)) %.%
mutate(cleansed_aDist = ApplyAdist(Company.Name.Compress.1,Company.Name.Compress.2)) %.%
mutate(cleansed_MaDist = ModifyAdist(Company.Name.Compress.1,Company.Name.Compress.2)) %.%
mutate(Clnsd_dist_over_len = cleansed_MaDist/NumChar)%.%
arrange(cleansed_MaDist)
results.fin$SD_MaD <- scale(results.fin$cleansed_MaDist)
results.fin$SD_DoL <- scale(results.fin$Clnsd_dist_over_len)